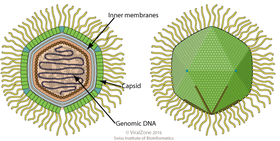
The typical form of the virions of the
Marseilleviridae is, in principle, similar to that of the
Mimiviridae.
Marseilleviridae is a family of viruses first named in 2012.[1] The genomes of these viruses are double-stranded DNA. Amoeba are often hosts, but there is evidence that they are found in humans as well.[2][3][4][5] The family contains one genus and four species, two of which are unassigned to a genus.[6][7] It is a member of the nucleocytoplasmic large DNA viruses clade.
Taxonomy
The genus contains the following genera and species:[7]
Related Viruses

Images of cryo-frozen
Marseilleviridae particles (left and center) and enlarged diagram of structure near a vertex. Black arrows indicate Large Dense Bodies. White arrows indicate lipid bilayer.
Additional species have since been recognized.[1] The first member of this family recognized has been named Acanthamoeba polyphaga marseillevirus. A second member is Acanthamoeba castellanii lausannevirus. Two additional viruses have been isolated but have yet to be named. Another member of this family has been isolated from blood donors.[4] An isolate from insects—Insectomime virus—has also been reported.[8]
The viruses appear to fall into at least 3 lineages: (1) Marseillevirus and Cannes8virus (2) Insectomime and Tunisvirus and (3) Lausannevirus. A sixth potential member of this family—Melbournevirus—appears to be related to the Marseillevirus/Cannes8virus clade.[9]
A seventh virus—Brazilian Marseillevirus—has been reported.[10] This virus appears to belong to a fourth lineage of virus in this family.
Another virus—Tokyovirus—has also been reported.[11]
Another member of this family is Kurlavirus.[12]
In 2017, it was proposed that the family contained the following five lineages:[13]
Lineage A
Lineage B
Lineage C
Lineage D
Lineage E
Another putative member of this family is Marseillevirus shanghai. If this virus is confirmed, it would belong to the A lineage.
Structure
Viruses in Marseilleviridae have icosahedral geometries. The diameter is around 250 nm. Genomes are circular, around 372kb in length. The genome has 457 open reading frames.[6]
Life cycle
DNA-templated transcription is the method of transcription. Amoeba serve as the natural host.[6]
Genomics
A promoter sequence—AAATATTT—has been found associated with 55% of the identified genes in this virus.[14] Most of these sequences occur in multiple copies.
History
One of the first members of this family was described in 2009.[15] Other members described around then (2007) and since then have been documented.[16]
References
-
^ a b Colson, Philippe; Pagnier, Isabelle; Yoosuf, Niyaz; Fournous, Ghislain; La Scola, Bernard; Raoult, Didier (2012). "'Marseilleviridae', a new family of giant viruses infecting amoebae". Archives of Virology. 158 (4): 915–20. doi:10.1007/s00705-012-1537-y. PMID 23188494.
-
^ La Scola, Bernard (2014). "Looking at protists as a source of pathogenic viruses". Microbial Pathogenesis. 77: 131–5. doi:10.1016/j.micpath.2014.09.005. PMID 25218687.
-
^ Colson, Philippe; Fancello, Laura; Gimenez, Gregory; Armougom, Fabrice; Desnues, Christelle; Fournous, Ghislain; Yoosuf, Niyaz; Million, Matthieu; La Scola, Bernard; Raoult, Didier (2013). "Evidence of the megavirome in humans". Journal of Clinical Virology. 57 (3): 191–200. doi:10.1016/j.jcv.2013.03.018. PMID 23664726.
-
^ a b Popgeorgiev, Nikolay; Boyer, Mickaël; Fancello, Laura; Monteil, Sonia; Robert, Catherine; Rivet, Romain; Nappez, Claude; Azza, Said; Chiaroni, Jacques; Raoult, Didier; Desnues, Christelle (2013). "Marseillevirus-Like Virus Recovered from Blood Donated by Asymptomatic Humans". The Journal of Infectious Diseases. 208 (7): 1042–50. doi:10.1093/infdis/jit292. PMID 23821720.
-
^ Aherfi, Sarah; Colson, Philippe; Audoly, Gilles; Nappez, Claude; Xerri, Luc; Valensi, Audrey; Million, Matthieu; Lepidi, Hubert; Costello, Regis; Raoult, Didier (2016). "Marseillevirus in lymphoma: A giant in the lymph node". The Lancet Infectious Diseases. 16 (10): e225–e234. doi:10.1016/S1473-3099(16)30051-2. PMID 27502174.
-
^ a b c "Viral Zone". ExPASy. Retrieved 12 August 2015.
-
^ a b "Virus Taxonomy: 2020 Release". International Committee on Taxonomy of Viruses (ICTV). March 2021. Retrieved 22 May 2021.
-
^ Boughalmi, Mondher; Pagnier, Isabelle; Aherfi, Sarah; Colson, Philippe; Raoult, Didier; La Scola, Bernard (2013). "First Isolation of a Marseillevirus in the Diptera Syrphidae Eristalis tenax". Intervirology. 56 (6): 386–94. doi:10.1159/000354560. PMID 24157885.
-
^ Doutre, G; Philippe, N; Abergel, C; Claverie, J.-M (2014). "Genome Analysis of the First Marseilleviridae Representative from Australia Indicates that Most of Its Genes Contribute to Virus Fitness". Journal of Virology. 88 (24): 14340–9. doi:10.1128/JVI.02414-14. PMC 4249118. PMID 25275139.
-
^ Dornas, Fábio; Assis, Felipe; Aherfi, Sarah; Arantes, Thalita; Abrahão, Jônatas; Colson, Philippe; La Scola, Bernard (2016). "A Brazilian Marseillevirus is the Founding Member of a Lineage in Family Marseilleviridae". Viruses. 8 (3): 76. doi:10.3390/v8030076. PMC 4810266. PMID 26978387.
-
^ Takemura, Masaharu (2016). "Draft Genome Sequence of Tokyovirus, a Member of the Family Marseilleviridae Isolated from the Arakawa River of Tokyo, Japan". Genome Announcements. 4 (3): e00429–16. doi:10.1128/genomeA.00429-16. PMC 4901213. PMID 27284144.
-
^ Chatterjee, Anirvan; Kondabagil, Kiran (2017). "Complete genome sequence of Kurlavirus, a novel member of the family Marseilleviridae isolated in Mumbai, India". Archives of Virology. 162 (10): 3243–3245. doi:10.1007/s00705-017-3469-z. PMID 28685284. S2CID 3984074.
-
^ Fabre E, Jeudy S, Santini S, Legendre M, Trauchessec M, Claverie J-M, et al (2017). Noumeavirus replication relies on a transient remote control of the host nucleus. Nat Commun 8:15087
-
^ Oliveira, Graziele Pereira; Lima, Maurício Teixeira; Arantes, Thalita Souza; Assis, Felipe Lopes; Rodrigues, Rodrigo Araújo Lima; Da Fonseca, Flávio Guimarães; Bonjardim, Cláudio Antônio; Kroon, Erna Geessien; Colson, Philippe; La Scola, Bernard; Abrahão, Jônatas Santos (2017). "The Investigation of Promoter Sequences of Marseilleviruses Highlights a Remarkable Abundance of the AAATATTT Motif in Intergenic Regions". Journal of Virology. 91 (21): e01088–17. doi:10.1128/JVI.01088-17. PMC 5640848. PMID 28794030.
-
^ Boyer, M; Yutin, N; Pagnier, I; Barrassi, L; Fournous, G; Espinosa, L; Robert, C; Azza, S; Sun, S; Rossmann, M. G; Suzan-Monti, M; La Scola, B; Koonin, E. V; Raoult, D (2009). "Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms". Proceedings of the National Academy of Sciences. 106 (51): 21848–53. Bibcode:2009PNAS..10621848B. doi:10.1073/pnas.0911354106. PMC 2799887. PMID 20007369.
-
^ Aherfi, Sarah; La Scola, Bernard; Pagnier, Isabelle; Raoult, Didier; Colson, Philippe (2014). "The expanding family Marseilleviridae". Virology. 466–467: 27–37. doi:10.1016/j.virol.2014.07.014. PMID 25104553.


 The typical form of the virions of the Marseilleviridae is, in principle, similar to that of the Mimiviridae.
The typical form of the virions of the Marseilleviridae is, in principle, similar to that of the Mimiviridae.