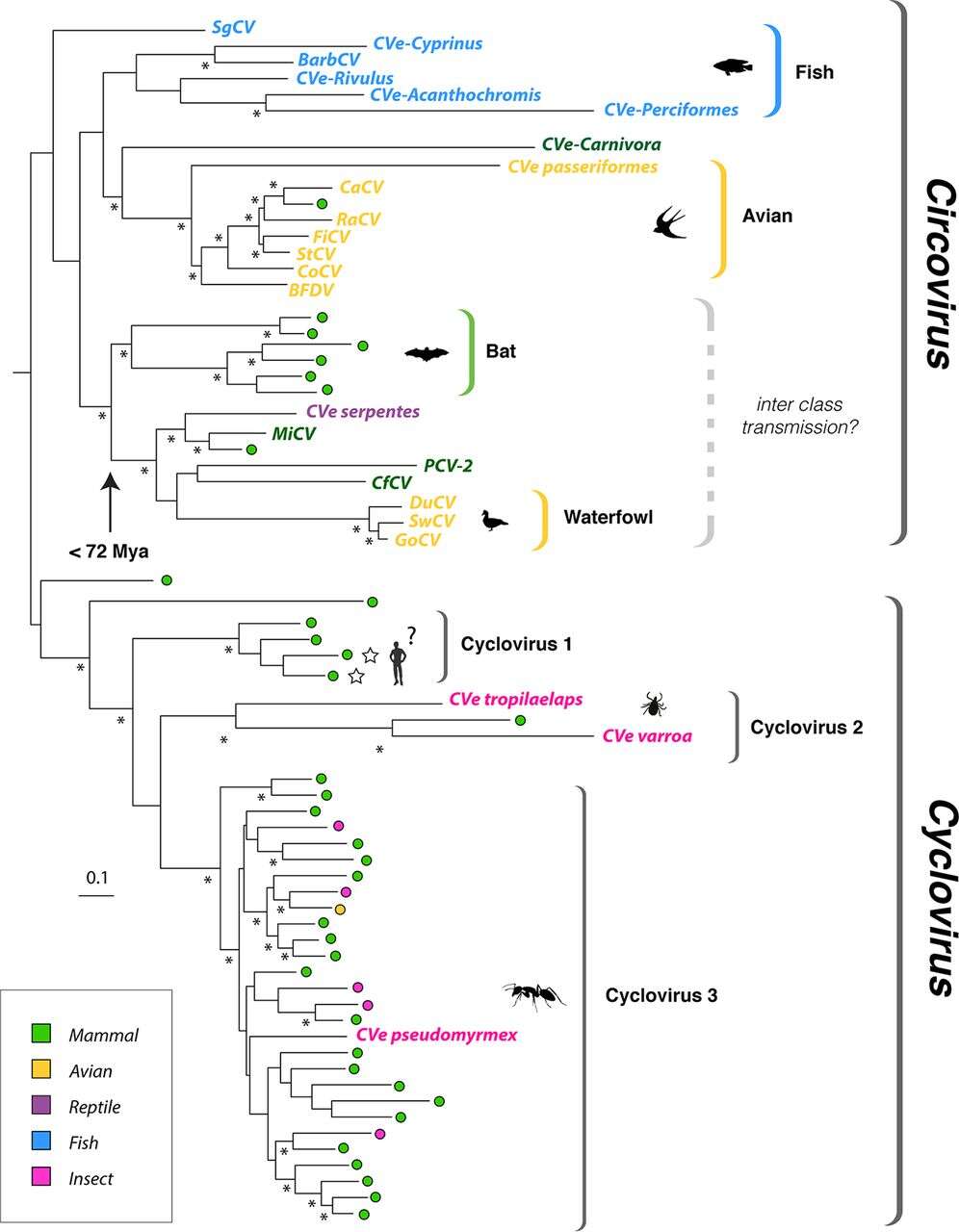
Description: English: Phylogeny of exogenous and endogenous circovirus Rep sequences. Maximum-likelihood phylogeny reconstructed from an alignment of replication-associated protein (Rep) sequences. The tree is midpoint rooted; asterisks indicate nodes with >70% bootstrap support. The scale bar indicates evolutionary distance in the number of substitutions per site. Sequences derived from metagenomic samples are indicated by colored circles. Taxon names are shown for sequences derived from viruses and CVe. All taxa are colored to indicate associations with host species groups, as shown in the key. Stars indicate viral taxa that have been linked to human disease. See Fig. S1 in the supplemental material for accession numbers of all taxa shown here. The arrow indicates an age calibration inferred for a clade within the Circovirus genus. Mya, million years ago; CaCV, canary circovirus; RaCV, raven circovirus; FiCV, finch circovirus; StCV, starling circovirus; CoCV, columbid circovirus; BFDV, beak and feather disease virus; MiCV, mink circovirus; PCV-2, porcine circovirus 2; CfCV, canine circovirus 1; DuCV, duck circovirus; SwCV, swan circovirus; GoCV, goose circoviruses; SgCV, wels catfish circovirus; BarbCV, barbel circovirus. Date: 31 July 2018. Source:
https://jvi.asm.org/content/jvi/92/16/e00145-18/F1.large.jpg?width=800&height=600&carousel=1 (Fig. 1) at
https://jvi.asm.org/content/92/16/e00145-18 Insights into Circovirus Host Range from the Genomic Fossil Record. ASM Journal of Virology 92 (16). e00145-18.
doi:10.1128/JVI.00145-18.
PMID 29875243.
. Author: Tristan P. W. Dennis, Peter J. Flynn, William Marciel de Souza, Joshua B. Singer, Corrie S. Moreau, Sam J. Wilson, Robert J. Gifford.