-
Locality: Spermonde archipelago, SW Sulawesi, Indonesia.
-
Euichi Hirose, Budhi Hascaryo Iskandar, Yusli Wardiatno
Zookeys
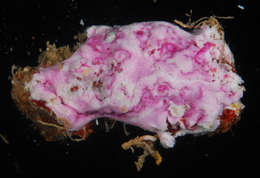
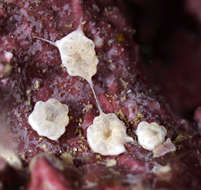
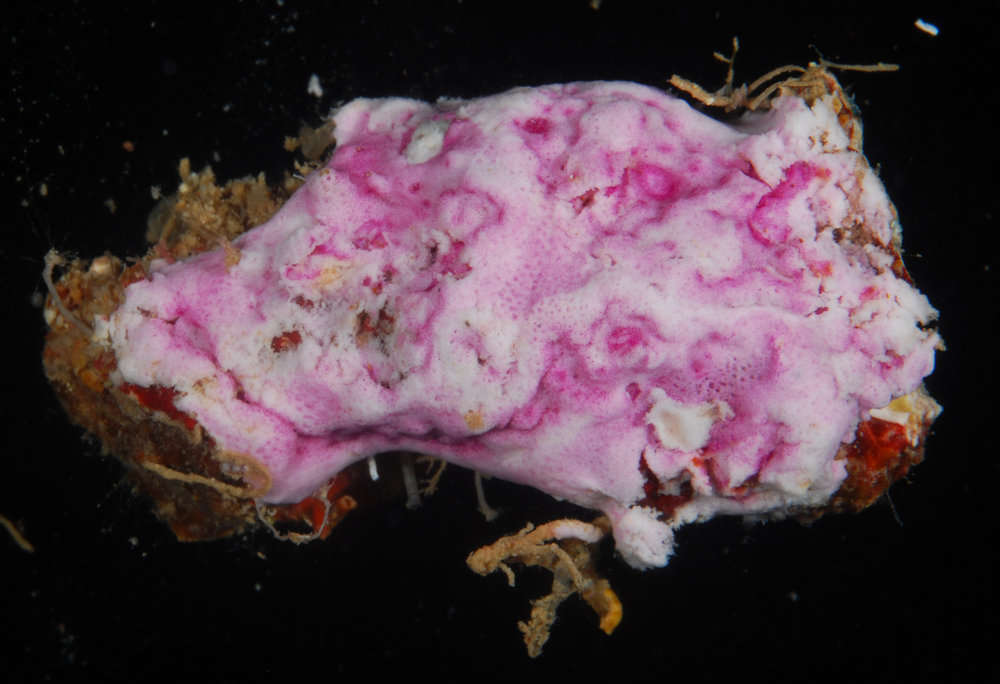
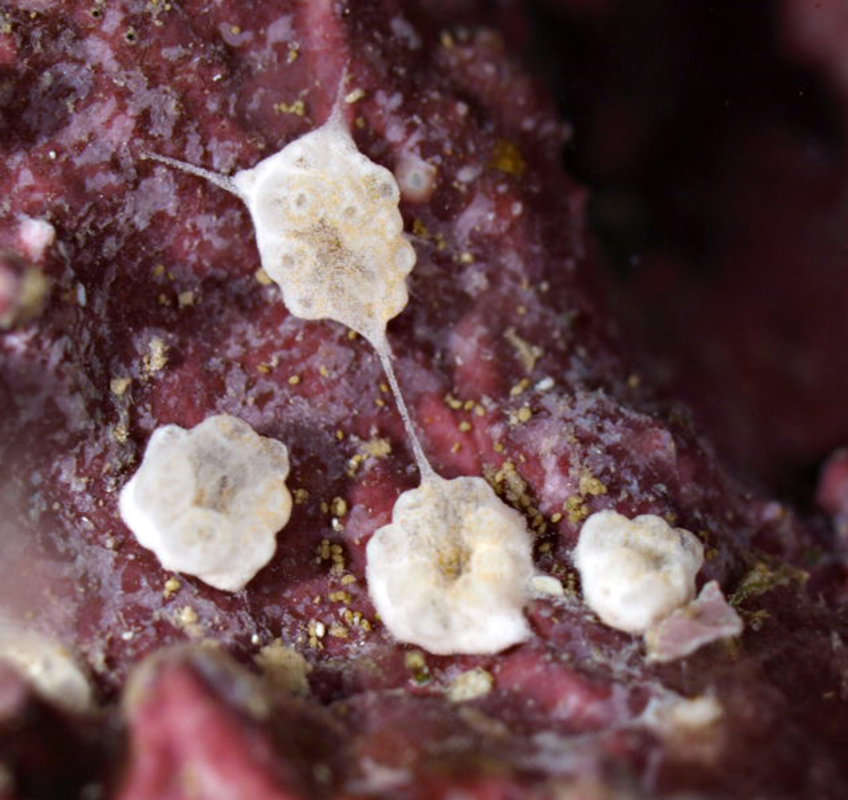
Figure 2.Photosymbiotic ascidians with tunic spicules. Colonies in situ and tunic spicules (inset) of Didemnum molle (A), Trididemnum miniatum (B), Lissoclinum patella (C), Lissoclinum punctatum (D), and Lissoclinum timorense (E). Tunic cells contain Prochloron cells in the tunic of Lissoclinum punctatum (F). Scale bars = 20 µm.
-
Locality: Spermonde archipelago, SW Sulawesi, Indonesia.
-
Euichi Hirose, Budhi Hascaryo Iskandar, Yusli Wardiatno
Zookeys
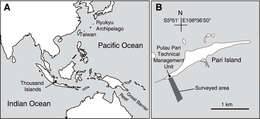
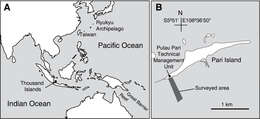
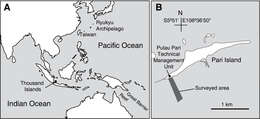
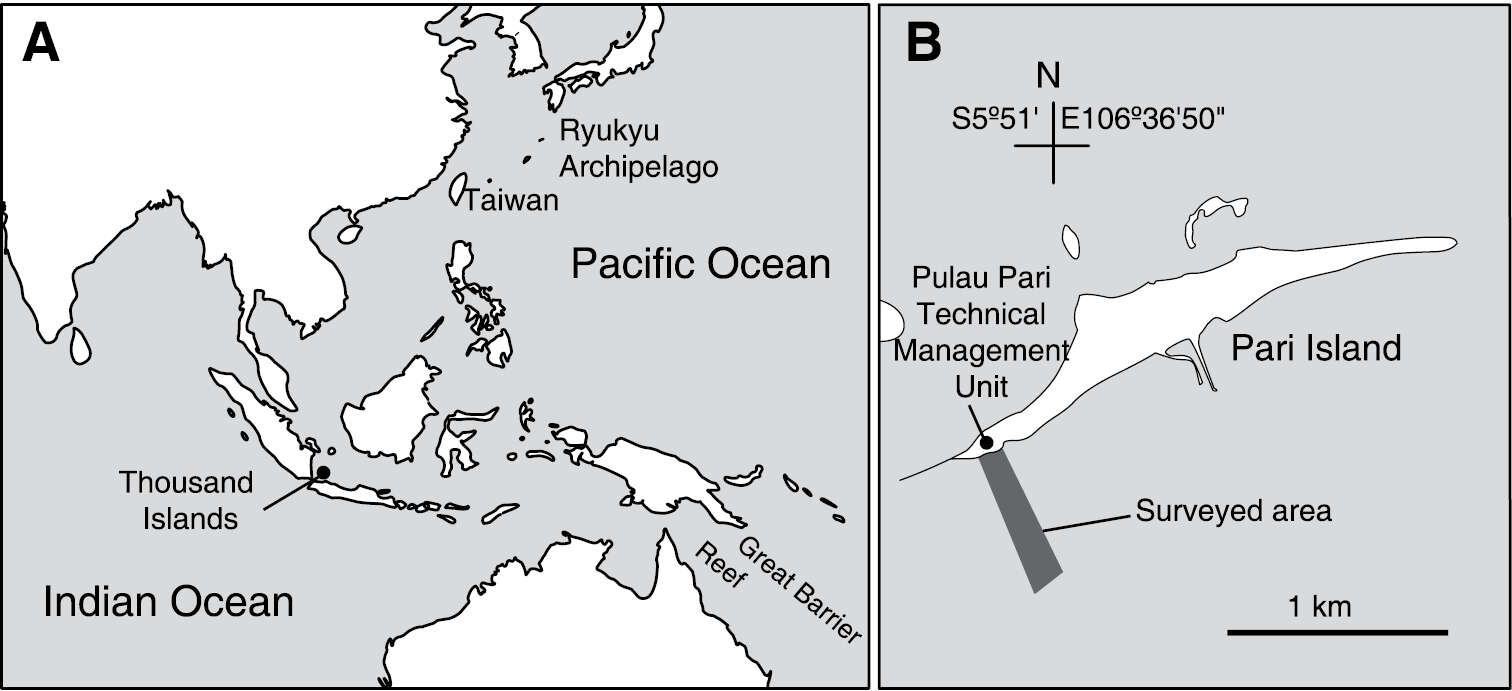
Figure 1.Location of Thousand Islands, Indonesia (A) and the surveyed area off Pari Island (B).
-
Euichi Hirose, Budhi Hascaryo Iskandar, Yusli Wardiatno
Zookeys
Figure 2.Photosymbiotic ascidians with tunic spicules. Colonies in situ and tunic spicules (inset) of Didemnum molle (A), Trididemnum miniatum (B), Lissoclinum patella (C), Lissoclinum punctatum (D), and Lissoclinum timorense (E). Tunic cells contain Prochloron cells in the tunic of Lissoclinum punctatum (F). Scale bars = 20 µm.
-
Euichi Hirose, Budhi Hascaryo Iskandar, Yusli Wardiatno
Zookeys
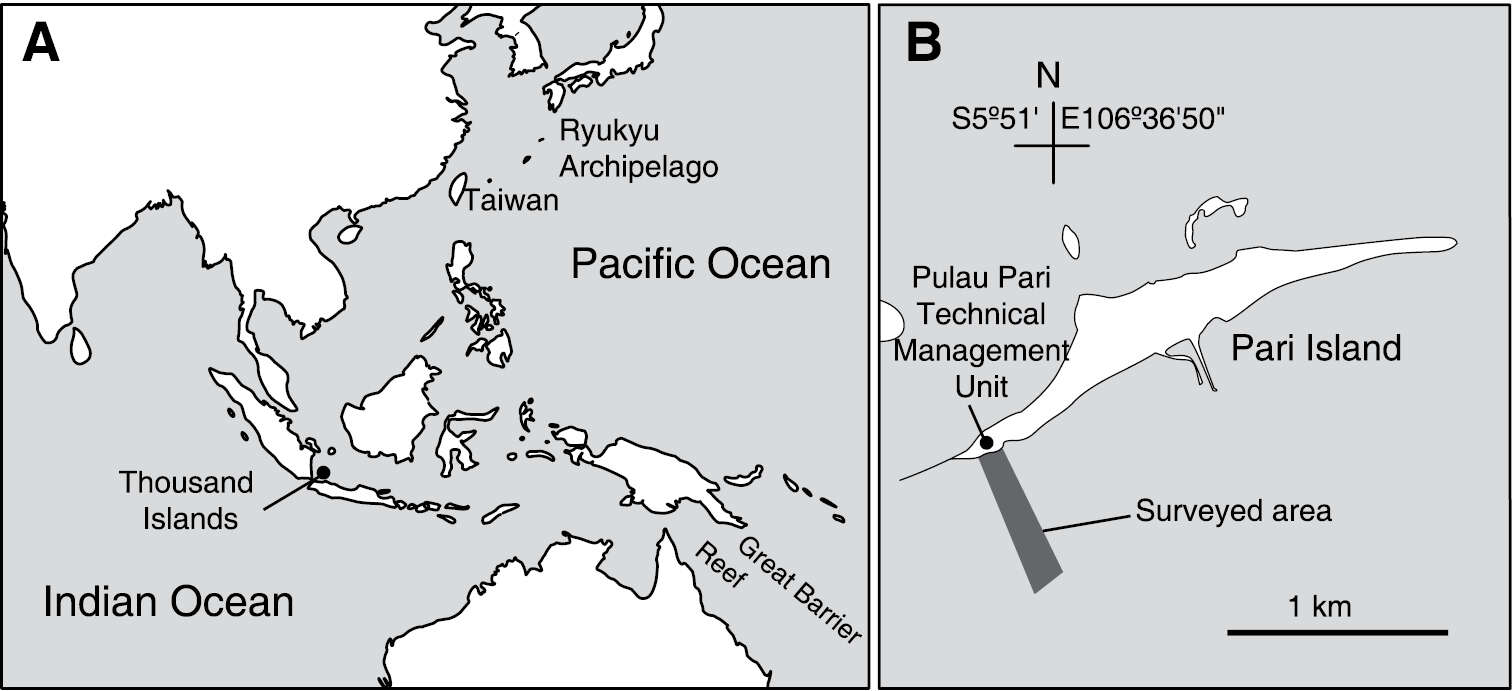
Figure 1.Location of Thousand Islands, Indonesia (A) and the surveyed area off Pari Island (B).
-
Euichi Hirose, Budhi Hascaryo Iskandar, Yusli Wardiatno
Zookeys
Figure 2.Photosymbiotic ascidians with tunic spicules. Colonies in situ and tunic spicules (inset) of Didemnum molle (A), Trididemnum miniatum (B), Lissoclinum patella (C), Lissoclinum punctatum (D), and Lissoclinum timorense (E). Tunic cells contain Prochloron cells in the tunic of Lissoclinum punctatum (F). Scale bars = 20 µm.
-
Euichi Hirose, Budhi Hascaryo Iskandar, Yusli Wardiatno
Zookeys
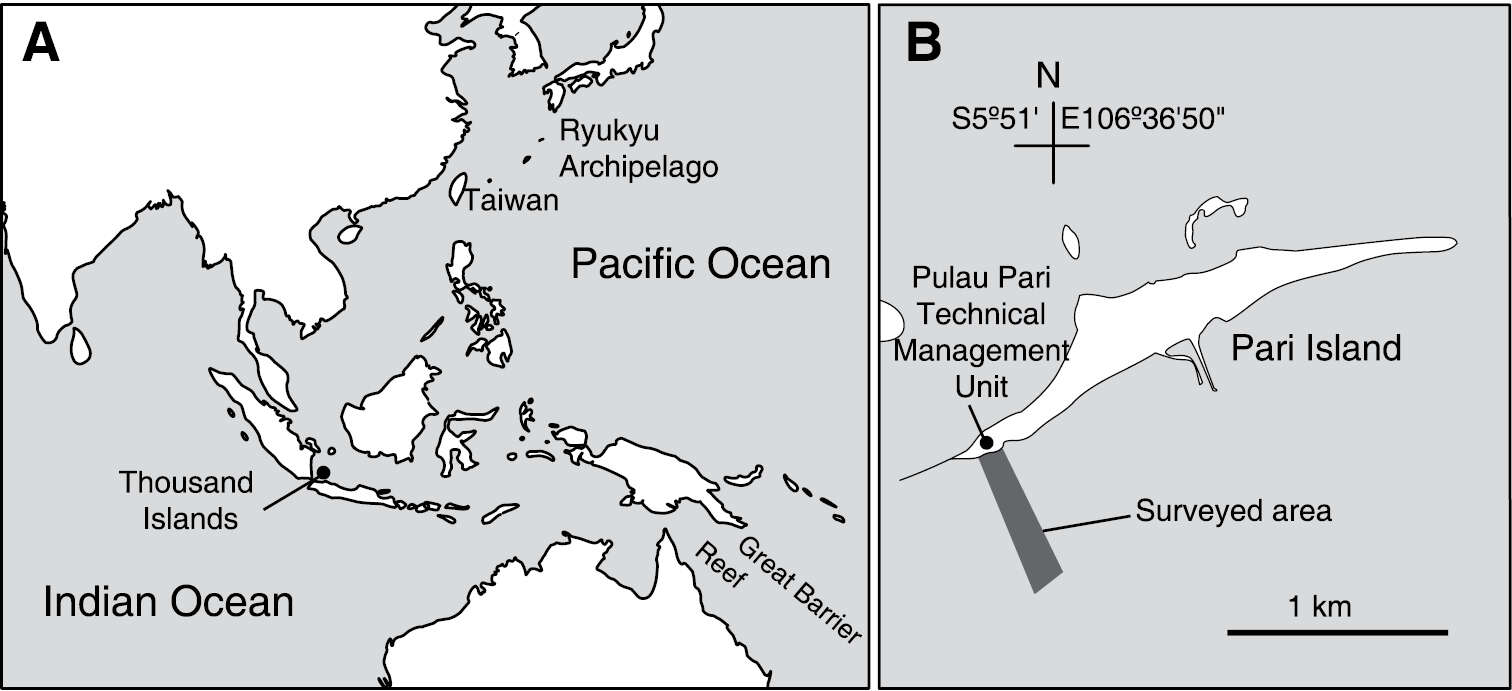
Figure 1.Location of Thousand Islands, Indonesia (A) and the surveyed area off Pari Island (B).
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
-
-
-
-
-
-