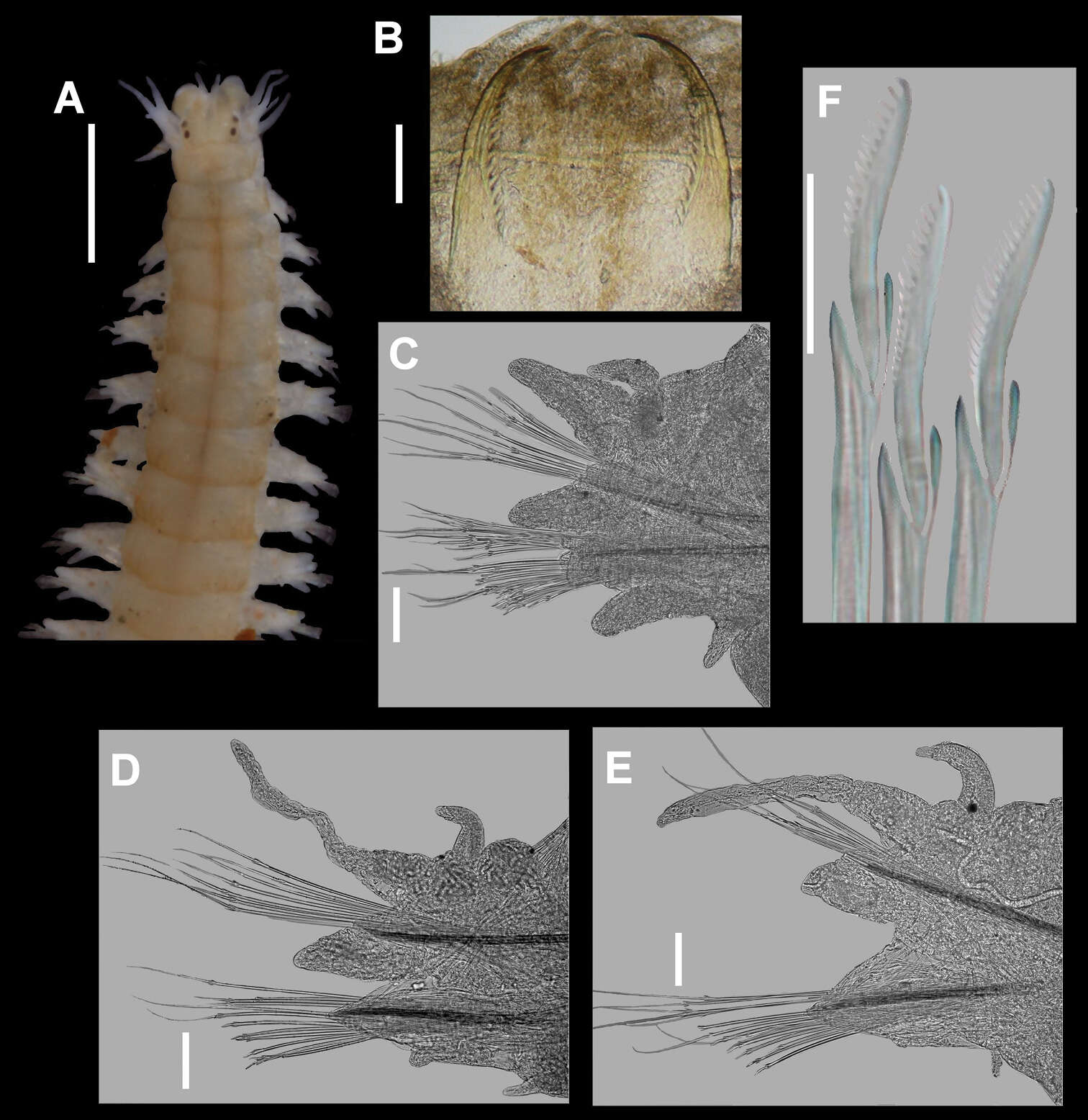
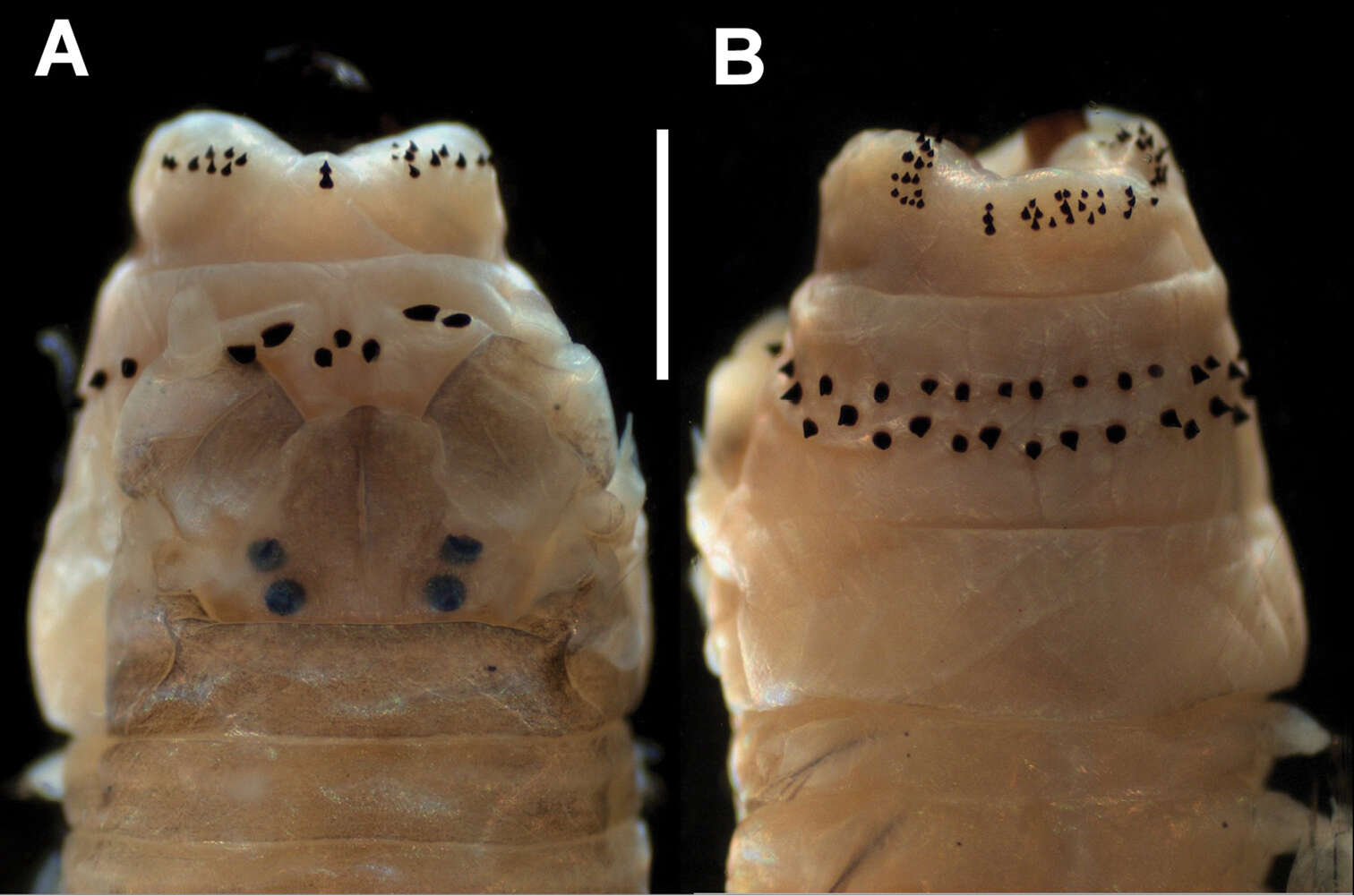
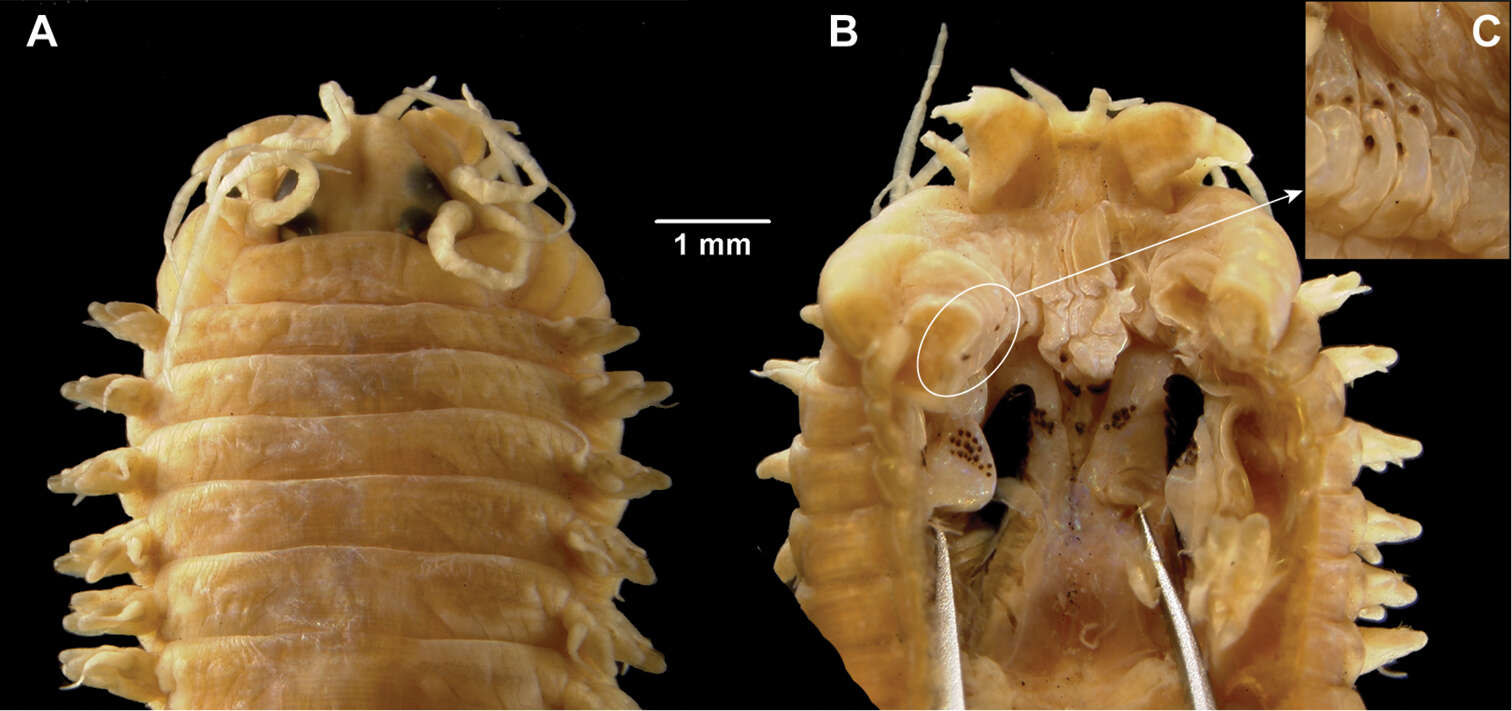
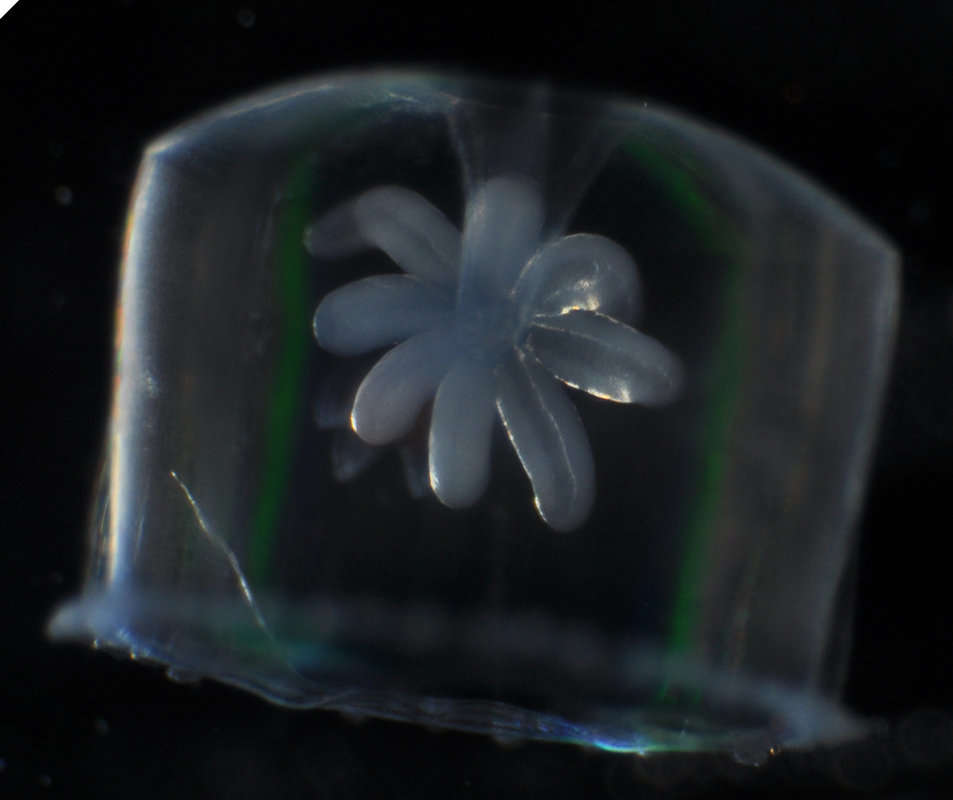
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
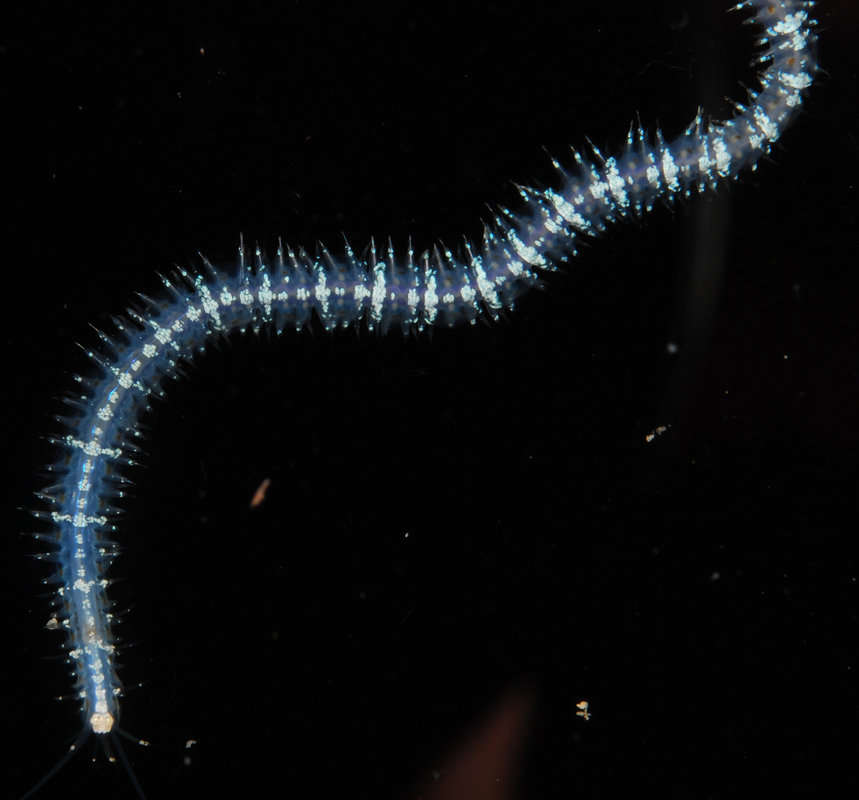
Description: The polychaete Nereis gracilis (Hansen, 1878). This motile species is known exclusively from deep
waters (range 930-2222 m) and has been recorded from the deeper parts of the Norwegian Sea, the Laptev Sea and
probably the Sea of Beaufort. As with many deep-sea species very little is known about its biology. Nereis gracilis is likely to be an omnivorous scavenger with the specimen from Dalsnuten being approximately 15 cm long. Item Type: Image Title: Nereis gracilis Copyright: SERPENT project Species: Nereis gracilis Behaviour: Motile and likely to be an omnivorous scavenger Site: Atlantic -- Norwegian -- Dalsnuten Site Description: Seafloor Depth (m): 1452 Latitude: 66 deg 34' 33" N Longitude: 3 deg 32' 46" E Countries: Norway -- Norwegian Sector Habitat: Benthic Rig: Aker Barents Project Partners: Shell, Aker Drilling, Oceaneering ROV: Magnum 142 Deposited By: Miss Moira MacLean Deposited On: 23 February 2011
-
Øresundsakvariet
-
Randers Fjord
-
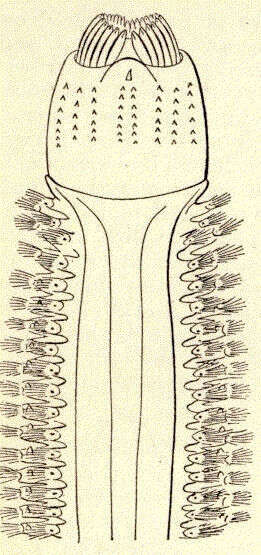
Nephthys ingens. Anterior part of body and extended proboscis; ventral view.
-
Glycera meekelii, with pharynx everted.
-
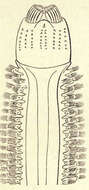
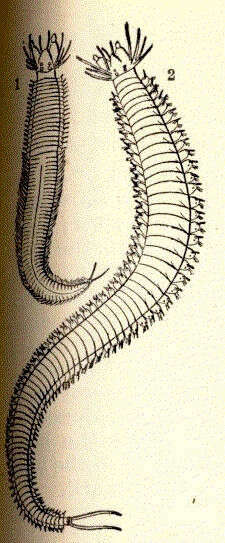
Nereis pelaglos. 1, male; 2, female.
-
Sarah Faulwetter, Georgios Chatzigeorgiou, Bella S. Galil, Artemis Nicolaidou, Christos Arvanitidis
Zookeys
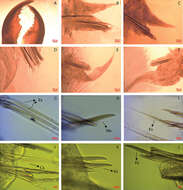
Figure 2.Sphaerosyllis levantina sp. n. SEM images of a anterior end and midbody, dorsal view b-c compound chaetae, anterior chaetigers d dorsalmost compound chaetae, anterior chaetiger e compound and dorsal simple chaetae, midbody f dorsalmost compound chaeta, posterior chaetiger g ventralmost compound chaetae, posterior chaetiger h dorsal simple chaeta
-
Mathan Magesh, Sebastian Kvist, Christopher J. Glasby
Zookeys
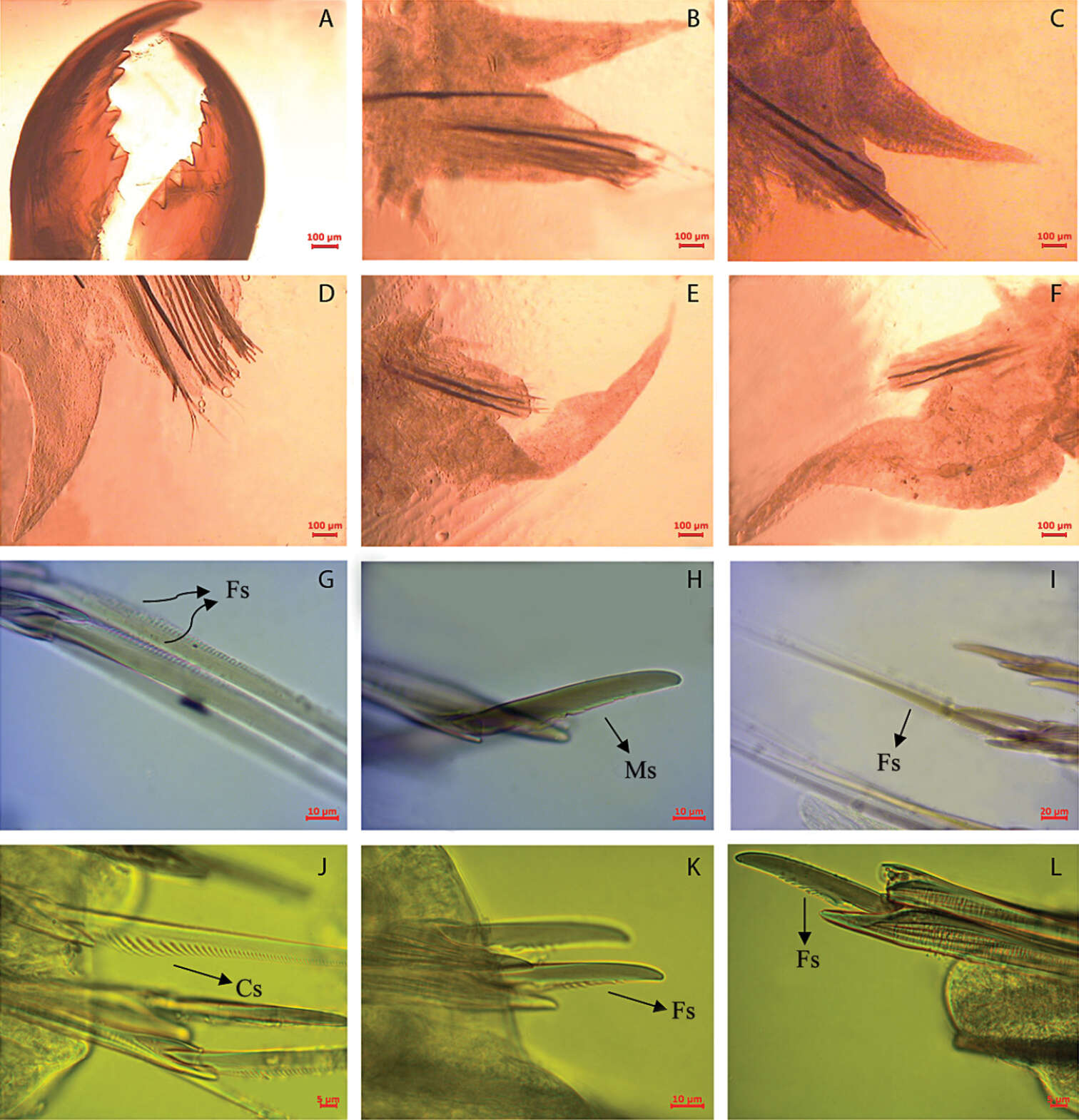
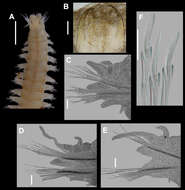
Figure 3.Namalycastis jaya sp. n.Holotype: a jaw pieces, ventromedial view b anterior parapodium from chaetiger 10 c parapodium from chaetiger 50 d parapodium from chaetiger 100 e posterior parapodium from chaetiger 210 f posterior parapodium from chaetiger 230 g sub-neuroacicular spiniger, chaetiger 10 h sub-neuroacicular falciger, chaetiger 109 i supra-neuroacicular spiniger, chaetiger 80 j sub-neuroacicular spiniger, chaetiger 210 k sub-neuroacicular falciger, chaetiger 210 l supra-neuroacicular falciger, chaetiger 20. Fs Finely serrated; Cs Coarsely serrated; Ms Medium serrated.
-
Jesús Angel de León-González, Berenice Trovant
Zookeys
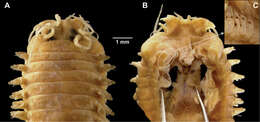
Figure 3.Nicon orensanzi sp. n. Paratype (UANL 7840). A Anterior end, dorsal view B Mandibles; Holotype (LACM) C Parapodium 9, anterior view D Parapodium 29, anterior view E Parapodium 62, anterior view F Infracicular sesquigomph falcigers of parapodium 62. Scale bars: A= 1 mm; B= 0.1 mm; C–E= 100 µ; F= 30µ.
-
Jesús Angel de León-González, Carrie A. Goethel
Zookeys
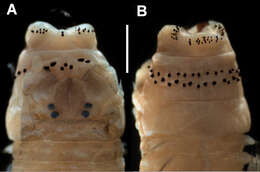
Figure 2.Perinereis rookeri sp. n. A Anterior end, dorsal view B Anterior end, ventral view. Measures: A–B = 1 mm.
-
Figure 6.Perinereis atlantica (McIntosh, 1885) (NHMUK.1885.12.1.161): A anterior end, dorsal view B anterior end, ventral view C enlarged view of partial Area VII–VIII of proboscis.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.